esemComp: a package for ESEM-within-CFA in R
The esemComp package
A while back I made a post about Exploratory Structural Equation Modeling (ESEM) in R. To be precise, how to do ESEM-within-CFA in R. Since then, I teamed up with Leon T. de Beer to develop esemComp, an R package to facilitate running these models. The package name is due to its main functionality allowing the composition of a model syntax to be run in the prominent SEM package lavaan.
Here I will run the same analysis as in my original post, but with esemComp functions. You’ll see that with the package the analysis gets much more straightforward. Furthermore, I’ll show how to do the analysis with a target rotation matrix for the exploratory factor analysis step, since this is the preferred approach. In this post, I’ll cut right to the chase, if you need a quick introduction to ESEM, check my original post.
We are going to use the toy dataset used in the examples by Morin, Marsh, and Nagengast (2013). It shows the data for 4500 participants in two occasions (3000 in the first, 1500 in the second). The scale is composed of six items, with three items related to a first factor and three to another, also, two of the items have significant cross-loadings. You can download the data here or at the original supplemental material.
Step one: EFA
First, load the data.
library(data.table) # for data wrangling
#if you need to install esemComp
# devtools::install_github("MateusPsi/esemComp")
library(esemComp)
esem_data <- fread("ESEM.dat")
# keep only first occasion and only the indicators
esem_data <- esem_data[V13 == 1, c(1:6)]
# rename variables to x1,x2...
names(esem_data) <- paste0("x", c(1:6)) | x1 | x2 | x3 | x4 | x5 | x6 |
|---|---|---|---|---|---|
| 0.10 | 0.30 | -0.42 | -0.54 | -1.79 | 1.64 |
| 1.69 | 1.95 | 2.41 | 2.53 | 2.53 | 0.84 |
| 0.71 | -0.17 | -1.25 | -0.49 | -1.65 | -0.48 |
| 2.09 | 0.37 | -0.14 | 1.27 | 0.67 | 2.72 |
| 0.03 | -0.15 | 1.22 | 0.06 | 0.90 | 1.09 |
Running the EFA with two factors and the GeominQ rotation in esemComp is actually pretty similar to using the psych package in R:
esem_efa <- esem_efa(esem_data, nfactors =2, fm = 'ml', delta = .5)
esem_efa$loadings##
## Loadings:
## ML1 ML2
## x1 0.827
## x2 0.696 0.226
## x3 0.525 0.313
## x4 0.138 0.762
## x5 0.160 0.672
## x6 0.554
##
## ML1 ML2
## SS loadings 1.491 1.489
## Proportion Var 0.249 0.248
## Cumulative Var 0.249 0.497If you want to use the approach recommended in some of the ESEM literature, a target rotation matrix, esemComp got you covered. A target rotation matrix is simply a matrix that represents an expected factor structure. The columns are the factors and the rows are the items, each cell is filled with either a zero or a one. In the rotation process, loadings corresponding to zeros are biased to be as close to zero as possible, and loadings corresponding to one are freely estimated.
In R, if we set ones, the values are actually biased toward one, so we use NA to represent free estimation. That is what the target matrix looks like for our toy data:
target_rot <- make_target(6,mainloadings = list(f1 = 1:3, f2 = 4:6))
target_rot## f1 f2
## [1,] NA 0
## [2,] NA 0
## [3,] NA 0
## [4,] 0 NA
## [5,] 0 NA
## [6,] 0 NAThen, we simply use the target rotation to run the EFA:
esem_target_efa <- esem_efa(esem_data, nfactors =2,
target = target_rot,
fm = 'ml')
esem_target_efa$loadings##
## Loadings:
## ML1 ML2
## x1 0.923 -0.200
## x2 0.751 0.103
## x3 0.551 0.230
## x4 0.781
## x5 0.110 0.681
## x6 -0.115 0.598
##
## ML1 ML2
## SS loadings 1.750 1.536
## Proportion Var 0.292 0.256
## Cumulative Var 0.292 0.548We can see that the target rotation makes the main loadings a little stronger and the cross-loadings a bit weaker. From here on, if we wanted to keep using the target rotation solution, we would only need to use this solution whenever a EFA object is needed in the next steps. We’ll continue to use the Geomin solution, tough, to be loyal to the orginial post.
Step two: set ESEM model
To compose the syntax of the ESEM-within-CFA model we need to identify a referent for each factor: an item that loads heavily on that factor but lightly on the others. We have a helper function for that in the package:
referents <- find_referents(esem_efa,factor_names = c("f1","f2"))Those referents make sense when we check the loadings from the EFAs.
Now to compose the model syntax to be used in lavaan. All loadings are estimated with the loadings of the EFA as starting points in ESEM-within-CFA, except for the crossloadings of the referent items.
Just pass the EFA object and the referents to the syntax composer and voilà!
#make model
esem_model <- syntax_composer(esem_efa, referents)
#print model
writeLines(esem_model)## f1 =~ start(0.827)*x1+
## start(0.696)*x2+
## start(0.525)*x3+
## 0.138*x4+
## start(0.16)*x5+
## start(-0.053)*x6
##
## f2 =~ -0.036*x1+
## start(0.226)*x2+
## start(0.313)*x3+
## start(0.762)*x4+
## start(0.672)*x5+
## start(0.554)*x6Step three: fit ESEM model
Finally, we fit the ESEM model to the data.
library(lavaan)
esem_fit <- cfa(esem_model, esem_data, std.lv=T)
summary(esem_fit, fit.measures = T, standardized = T)## lavaan 0.6-8 ended normally after 23 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 17
##
## Number of observations 3000
##
## Model Test User Model:
##
## Test statistic 3.897
## Degrees of freedom 4
## P-value (Chi-square) 0.420
##
## Model Test Baseline Model:
##
## Test statistic 6628.732
## Degrees of freedom 15
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 1.000
## Tucker-Lewis Index (TLI) 1.000
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -21698.913
## Loglikelihood unrestricted model (H1) -21696.964
##
## Akaike (AIC) 43431.825
## Bayesian (BIC) 43533.933
## Sample-size adjusted Bayesian (BIC) 43479.918
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.000
## 90 Percent confidence interval - lower 0.000
## 90 Percent confidence interval - upper 0.027
## P-value RMSEA <= 0.05 1.000
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.004
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|)
## f1 =~
## x1 0.885 0.022 40.638 0.000
## x2 0.636 0.022 28.634 0.000
## x3 0.526 0.022 24.210 0.000
## x4 0.138
## x5 0.125 0.020 6.124 0.000
## x6 -0.055 0.022 -2.560 0.010
## f2 =~
## x1 -0.036
## x2 0.212 0.023 9.115 0.000
## x3 0.323 0.023 13.906 0.000
## x4 0.883 0.021 42.265 0.000
## x5 0.585 0.020 28.677 0.000
## x6 0.473 0.021 22.414 0.000
## Std.lv Std.all
##
## 0.885 0.827
## 0.636 0.690
## 0.526 0.517
## 0.138 0.120
## 0.125 0.145
## -0.055 -0.065
##
## -0.036 -0.034
## 0.212 0.230
## 0.323 0.318
## 0.883 0.770
## 0.585 0.679
## 0.473 0.560
##
## Covariances:
## Estimate Std.Err z-value P(>|z|)
## f1 ~~
## f2 0.448 0.028 16.025 0.000
## Std.lv Std.all
##
## 0.448 0.448
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .x1 0.390 0.028 13.948 0.000
## .x2 0.279 0.014 20.369 0.000
## .x3 0.501 0.015 32.595 0.000
## .x4 0.407 0.024 16.625 0.000
## .x5 0.320 0.013 25.007 0.000
## .x6 0.510 0.015 32.923 0.000
## f1 1.000
## f2 1.000
## Std.lv Std.all
## 0.390 0.340
## 0.279 0.328
## 0.501 0.485
## 0.407 0.309
## 0.320 0.430
## 0.510 0.715
## 1.000 1.000
## 1.000 1.000Quick figure with semPlot:
library(semPlot)
semPaths(esem_fit,"std")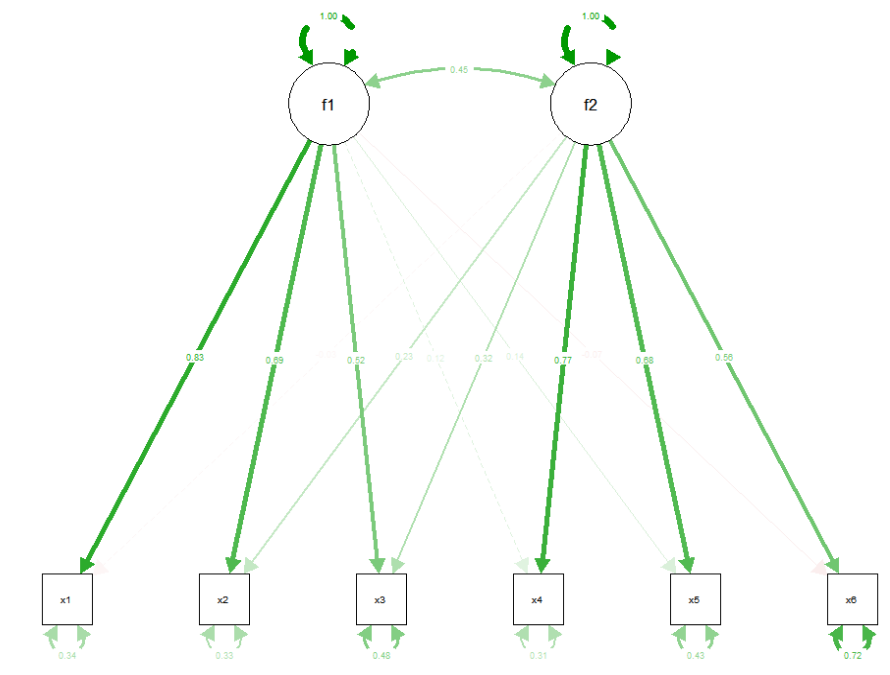
And that is it, ESEM-within-CFA made simple!